| Citation: | Yingqiu Zheng, Huachen Liu, Xin Dang, Juan Diego Gaitán-Espitia, Muyan Chen. 2025. Functional evolution of thyrotropin-releasing hormone neuropeptides: Insights from an echinoderm. Zoological Research, 46(1): 236-248. DOI: 10.24272/j.issn.2095-8137.2024.256 |
Feeding behavior is regulated by a complex network of endogenous neuropeptides. In chordates, this role is suggested to be under the control of diverse factors including thyrotropin-releasing hormone (TRH). However, whether this regulatory activity of TRH is functionally conserved in non-chordate metazoans, and to what extent this process is underpinned by interactions of TRH with other neuropeptides such as cholecystokinin (CCK, known as a satiety signal), remain unclear. This study investigated the TRH signaling system in the echinoderm Apostichopus japonicus. Bioinformatic analyses and ligand-binding assays identified a functional TRH receptor (AjTRHR) that activated signaling via the MAPK/ERK1/2 pathways. Experimental administration of TRH significantly reduced feeding activity, while up-regulating CCK expression. RNA interference (RNAi) experiments confirmed that both CCK and TRH are essential components of satiety signaling, working synergistically to mediate feeding inhibition. Evolutionary analysis of TRH-type peptides revealed greater conservation of the short isoform of TRH compared to the long isoform, probably driven by strong selection acting on the functional redundancy. These findings provide compelling evidence of a TRH-mediated signaling system in non-chordate deuterostomes, expanding our understanding of neuropeptide-regulated feeding mechanisms in marine invertebrates.
Feeding is a pivotal behavior in animal growth, development, reproduction, energetic homeostasis, and immune function. This intricate process is governed by complex neural circuits (Guo et al., 2021; Pool & Scott, 2014) that coordinate appetite and satiety signals through the actions of neuropeptides (Root et al., 2011). These signaling molecules, fundamental components of the neuroendocrine system, play critical roles in regulating diverse physiological and behavioral processes in animals (Burbach, 2011). Notably, neuropeptides, such as thyrotropin-releasing hormone (TRH), have been widely implicated as key regulators of feeding behavior (Al-Arabi & Andrews, 2006; Puga et al., 2016; Schuhler et al., 2007; Schwartz et al., 2013; Steward et al., 2003). Most of our current knowledge regarding the functional role of TRH in feeding regulation is derived from vertebrate studies. In rodents, such as rats and squirrels, TRH administration has been shown to suppress feeding motivation and inhibit feeding behavior (Al-Arabi & Andrews, 2006; Frare et al., 2021; Puga et al., 2016; Schuhler et al., 2007; Schwartz et al., 2013; Steward et al., 2003). However, the extent to which these effects are conserved across other taxa, particularly in relation to the evolutionary history of TRH signaling, remains unclear. Given the estimated 560-million-year evolutionary trajectory of this neuropeptide signaling pathway, its functional role in diverse chordate lineages and beyond is still not fully understood. This uncertainty is further compounded by the identification of TRH-type neuropeptides in various invertebrate metazoans, including mollusks (fulicins) (Ohta et al., 1991) and annelids/arthropods/nematodes/priapulids (EFLGamides) (Bauknecht & Jékely, 2015; Conzelmann et al., 2013; Van Sinay et al., 2017; Veenstra & Šimo, 2020; Zandawala et al., 2024), and in non-vertebrate deuterostomes (TRH) (Chen et al., 2019; Jékely, 2013; Rowe & Elphick, 2012; Semmens et al., 2016; Zandawala et al., 2017). Despite this broad taxonomic distribution, the functional roles of TRH-like neuropeptides in feeding regulation within these invertebrate groups remain largely uncharacterized.
Echinoderms, as deuterostome invertebrates, share a close evolutionary relationship with chordates, forming a sister group alongside hemichordates. This phylogenetic position, combined with ecological traits shared with marine protostomes (e.g., mollusks, annelids, and arthropods), such as feeding strategies, external fertilization, and larval development, provides an interesting opportunity to study and compare the evolutionary and functional conservationism of TRH and other neuropeptides in the regulation of feeding behavior. TRH-type neuropeptides have been identified across diverse echinoderm classes, including sea urchins (e.g., Strongylocentrotus purpuratus) (Rowe & Elphick, 2012), starfish (e.g., Asterias rubens and Acanthaster planci) (Semmens et al., 2016; Smith et al., 2017), ophiuroids (e.g., Ophionotus victoriae, Amphiura filiformis, and Ophiopsila aranea) (Zandawala et al., 2017), and sea cucumbers (e.g., Apostichopus japonicus, Holothuria scabra, Holothuria glaberrima, and Holothuria leucospilota) (Chen et al., 2019; Chieu et al., 2019; Rowe et al., 2014; Suwansa-Ard et al., 2018). Empirical evidence suggests that TRH-type neuropeptides in echinoderms are implicated in critical processes, including gonadal development in adults and feeding, attachment, and metamorphosis of larvae (Chaiyamoon et al., 2020; Mayorova et al., 2016; Zheng et al., 2022). Previous studies have uncovered key insights into the role of TRH-type neuropeptides in sea cucumbers, particularly their involvement in the up-regulation of the TRH-type precursor AjTRHP during aestivation, a physiological state during which feeding is suspended (Zhao et al., 2014). Recent single-cell RNA sequencing in A. japonicus also identified a neuronal population with markedly elevated AjTRHP expression, with TRH predicted to function as the central regulatory gene within this neuronal network governing feeding behavior (Zheng et al., 2024). Despite these advances, the precise mechanisms through which TRH regulates feeding cessation in sea cucumbers, and its potential parallels with vertebrate deuterostomes, remain unresolved, raising questions about the functional and evolutionary conservation of TRH-mediated feeding regulation (Zuckerkandl & Pauling, 1965).
To address these questions, the present study employed an integrated approach combining cellular, biochemical, and molecular methodologies to reconstruct the structure and dynamics of the TRH signaling pathway in A. japonicus. Furthermore, interactions between TRH and other neuropeptides potentially involved in satiety signaling, such as cholecystokinin (CCK), were explored. Notably, our study revealed the function of the TRH signaling system in feeding regulation within a non-chordate deuterostome, offering new insights into the neuroendocrine mechanisms underlying feeding regulation in marine invertebrates.
Experimental animals were obtained from the Shandong Oriental Ocean Sea Cucumber Breeding Farm (Weihai, China) and transferred to the aquarium facilities at Ocean University of China. Laboratory culture conditions utilized filtered and ultraviolet (UV)-sterilized seawater sourced from the Qingdao coastal region, maintained at a temperature of 18–20°C, salinity of 32 ppt, and 12:12 h light/dark cycle. Following acclimation for one week, tissues including the circumoral nerve ring (CNR), intestine, longitudinal muscle, and gonads (male/female) were collected from six adults (106±3.82 g). Samples were snap-frozen in liquid nitrogen and stored at −80°C for reverse transcription and quantitative real-time PCR (RT-qPCR) analysis. The CNR and intestinal tissues were also fixed in 4% paraformaldehyde (PFA) in 0.1 mol/L phosphate-buffered saline (PBS) (G-CLONE, China) for in situ hybridization (ISH).
For injection experiments targeting AjTRH1 and AjTRH7, 180 sub-adult sea cucumbers (30±5.8 g) were randomly divided into six groups (30 individuals per group). Animals in each group were then transferred to five fiber-glass tanks (six individuals per tank). Samples for qPCR analysis of AjTRHP expression in the CNR and intestine were obtained from one individual per tank. For RNA interference (RNAi) experiments, 30 sea cucumbers (30±3.01 g) were randomly assigned to three groups (six individuals per group). During the experimental period, sea cucumbers were fed ad libitum each morning with a diet comprising a mixture of artificial commercial food and sea mud (1:3 ratio). The artificial food, purchased from the Shandong Oriental Ocean Sea Cucumber Breeding Farm (Weihai, China), included a blend of kelp, scallop skirt, brown alga (Sargassum thunbergii), gulfweed, Ulva, and yeast. Each tank was provided with 40–50 g of the diet daily, and uneaten food and feces were removed before the subsequent feeding to maintain optimal water quality.
The full-length cDNA sequence of AjTRHP was reported in our previous work (Zheng et al., 2022). Gene structure was predicted based on the A. japonicus reference genome (China National GeneBank DataBase (CNGBdb) under BioProject accession No. CNP0002776), and conserved domains were analyzed using the Simple Modular Architecture Research Tool (SMART). Synteny analysis was conducted by identifying the chromosomal locations of TRHP in the genomes of Apostichopus japonicus, Asterias rubens, and S. purpuratus, using publicly available data from NCBI (https://www.ncbi.nlm.nih.gov/). The three-dimensional (3D) structure of AjTRHP protein was predicted using the Robetta online platform (https://robetta.bakerlab.org/). Sequence alignments of mature TRH-type peptides from A. japonicus and other bilaterians (GenBank IDs, Supplementary Table S1) were performed using ClustalW in MEGA7 (v.7170509) with default parameters.
Total RNA was extracted from tissues, including the CNR, intestine, longitudinal muscle, and gonads (male/female), using TRIzol reagent (Takara, Japan). RNA quality and integrity were determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) and 1% agarose gel electrophoresis (OD260/280=1.9–2.1, c>30 ng/µL, 28S/18S≥1.0). Relative gene expression levels were quantified using TB Green® Premix Ex Taq™ (Tli RnaseH Plus) (Takara, Japan) on a StepOnePlus Real-Time PCR System (ABI, USA). All reactions were conducted in triplicate. β-actin and β-tubulin were used as reference genes for normalization, and relative expression levels were analyzed using the 2−ΔΔCT method (significance threshold alpha=0.05), as described previously (Zheng et al., 2022). Results are presented as mean±standard error of the mean (SEM) (n=6). The specific primers for AjTRHP amplification are given in Supplementary Table S2.
TRHR from the sea urchin S. purpuratus (GenBank accession No. XM_011684235.1) was used as a reference to identify candidate TRH-type neuropeptide receptors in the A. japonicus chromosome-level genome and CNR transcriptome data (GenBank accession No. GHCH00000000). Among the identified sequences, the top-scoring transcript (GenBank accession No. PIK53909.1) was designated as the candidate A. japonicus TRH receptor (AjTRHR). Full-length cDNA sequences of AjTRHR were then cloned using a SMARTer® RACE 5’/3’ Kit (Clontech, Japan) with specific primers (Supplementary Table S2) following the manufacturer’s instructions.
A three-dimensional (3D) model of the AjTRHR protein was modeled using SWISS-MODEL (https://swissmodel.expasy.org/), while its transmembrane domain (TMD) was predicted using Protter (http://wlab.ethz.ch/protter/start/). To explore the evolutionary relationships between AjTRHR and TRHR-like protein in other species, a phylogenetic analysis was performed using the maximum-likelihood (ML) method implemented in MEGA7 (v.7170509) using default parameters. After trimming the leading and trailing sequence ends, the General Reversible Chloroplast+Frequency model (cpREV+G+I+F) was selected as the optimal substitution model. The analysis included 1 000 bootstrap replicates to infer a robust consensus tree. Receptors for gonadotropin-releasing hormone (GnRH)-like proteins, known to be closely related to TRH, were also included in this analysis (Suwansa-Ard et al., 2018). Kisspeptin receptor (KissR)-like sequence was selected as the outgroup for phylogenetic analysis. GenBank IDs of all sequences are listed in Supplementary Table S1. The alignment sequence file (Supplementary File 1) and Newick tree file (Supplementary File 2) are provided in the Supplementary Materials.
AjTRH1 (pQYFA, the short peptide isoform), AjTRH7 (pQLPGSPWKFWE, the long peptide isoform) along with their reverse sequences (AFYQ and EWFKWPSGPLQ), were synthesized by GL Biochem (Shanghai) Ltd. (China). The open reading frame (ORF) sequences of AjTRHR were subcloned into the pcDNA 3.1(+) and pEGFP-N1 vectors (Biofeng, China) to generate endo-free plasmids for transfection, as described previously (Li et al., 2022). Primer sequences used for cloning are provided in Supplementary Table S2.
Ligand binding assays, including intracellular calcium measurements and receptor localization and translocation assay were performed to confirm the ligand activity of AjTRH1 and AjTRH7 for AjTRHR. Human embryonic kidney (HEK293T) cells were purchased from ATCC (USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM, HyClone, USA) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Israel) at 37°C in a 5% CO2-humidified atmosphere. The HEK293T cells were plated on 6-well plates (Corning, USA) and transfected with AjTRHR plasmids using Lipofectamine 2000 Reagent (Invitrogen, USA) at approximately 70% confluence according to the manufacturer’s instructions.
Intracellular calcium flux was measured using the fluorescent Ca2+ indicator Fura-2/AM and SRE-Luc reporter assays (Li et al., 2010). Briefly, HEK293T cells expressing AjTRHR-pcDNA 3.1(+) were incubated with 3 μmol/L Fura-2/AM (Thermo Fisher Scientific, USA) for 30 min and stimulated with AjTRH1/AjTRH7 at indicated concentrations (10−9–10−5 mol/L). Calcium flux was detected using a Synergy H1 Hybrid Multi-Mode Reader (Biotek, USA). Receptor localization and translocation assays were conducted as described in previous research (Wang et al., 2020). HEK293T cells expressing AjTRHR-pEGFP-N1 were seeded on glass coverslips in 6-well plates and stimulated with 1 μmol/L AjTRH1/AjTRH7 for 30 min. The cells were fixed with 4% PFA in PBS at room temperature for 10 min, incubated with 4’,6-diamidino-2-phenylindole (DAPI, Beyotime, China) for 5 min, and visualized using an Olympus BX53F fluorescence microscope system (Japan). All ligand-binding experiments were repeated independently at least three times. Negative control groups included cells transfected with empty pcDNA3.1 (+) or pEGFP-N1 vectors or stimulated with reverse peptide sequences.
HEK293T cells co-transfected with AjTRHR/pcDNA 3.1(+) and pCRE-Luc were used for cAMP-response element (CRE)-Luciferase (Luc) reporter gene assays. Cells were seeded into 12-well cell culture plates and incubated with AjTRH1/AjTRH7 (10−9–10−5 mol/L) for 4 h at 37°C. Luciferase activity was detected using a Firefly Luciferase Reporter Gene Assay Kit (MKBio, China) following the manufacturer’s instructions. The cAMP assay was performed as described previously (Wang et al., 2017). Briefly, gradient-diluted AjTRH1/AjTRH7 (10−9–10−5 mol/L) was used to treat AjTRHR-pcDNA3.1 (+) or pcDNA3.1 (+) transfected cells for 30 min. After treatment, the cells were washed twice with PBS, lysed, and collected. Intracellular cAMP levels were quantified using a cAMP Assay Kit (R&D Systems, USA) following the manufacturer’s protocols. All data were processed and imaged using GraphPad Prism v.9.4 (GraphPad Software, USA).
Phosphorylated ERK1/2 (pERK1/2) activity was measured as described in previous research (Wang et al., 2020). Briefly, cells expressing AjTRHR-pcDNA 3.1(+) were treated with AjTRH7 (10−6 mol/L) for varying durations (0, 5, 15, 30, 45, and 60 min) at 37°C. After treatment, cells were lysed, and total protein concentrations were determined using an Enhanced BCA Protein Assay Kit (Beyotime, China). Protein samples were separated using 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes. Immunoblotting was performed using the primary antibodies rabbit anti-pERK1/2 (1:2 000, Cell Signaling Technology, USA) and rabbit anti-tERK1/2 (1:1 000, Cell Signaling Technology, USA). A goat anti-rabbit IgG secondary antibody (1:2 000, Absin, China) was used for detection. Immunoreactive bands were visualized using an enhanced chemiluminescent reagent (Beyotime, China) and quantified using ImageJ. Negative controls included cells transfected with empty vectors.
ISH experiments were performed to localize the transcripts of AjTRHP and AjTRHR, following established protocols (Li et al., 2022). Specific AjTRHP and AjTRHR antisense and sense digoxygenin (DIG)-labeled RNA probes were synthesized using a DIG RNA Labelling Kit (Roche, Switzerland). The primer sequences used for probe generation are listed in Supplementary Table S2. Tissues from A. japonicus were fixed in 4% PFA prepared in 0.1 mol/L PBS for 12 h at 4°C. Following fixation, tissues were embedded in paraffin, sectioned, and processed through dewaxing, permeabilization, and proteinase K digestion. After prehybridization and probe hybridization, the sections were stained using a DIG Nucleic Acid Detection Kit (Roche, Switzerland) and visualized using an Olympus bright field light microscope (Japan).
To investigate the physiological roles of AjTRH1 and AjTRH7 in A. japonicus, in vivo pharmacological experiments were performed via AjTRH1/AjTRH7 injection into the coelom. Based on previous studies of neuropeptide effects in A. japonicus and the reported activity of TRH peptides in other species, optimal injection concentrations were set to 0.5 mg/mL or 0.05 mg/mL (Wang et al., 2020). AjTRH1 and AjTRH7 were dissolved in sterile seawater to a final concentration of 0.5 mg/mL or 0.05 mg/mL and administered via the oral cavity directly into the coelom every two days. A total of 180 sea cucumbers were randomly divided into six defined groups: high-concentration AjTRH1/AjTRH7 (0.5 mg/mL) injection groups (HTG), low-concentration AjTRH1/AjTRH7 (0.05 mg/mL) injection groups (LTG), and two seawater injection groups (NC). Each injection volume was set at 0.1% (v/w) and administered once every two days at noon. Sea cucumbers were maintained under identical feeding conditions throughout an over 25-day experimental period. Uneaten food was collected daily, dried, and weighed to monitor feed intake. The body weight of each individual was measured at the beginning and end of the experiment. Following the treatment period, fresh CNR and intestinal tissues were dissected and frozen in liquid nitrogen. The weight gain rate (WGR) and feed intake rate (FI) of 30 individuals per treatment were calculated as described previously (Liu et al., 2014).
Knockdown experiments targeting AjTRHP were performed in vivo as described previously (Huang et al., 2021). Two specific siRNA sequences targeting AjTRHP (siRNA_AjTRHP) were designed and synthesized by GenePharma (China), along with a negative control (siRNA_NC), which was confirmed not to target any annotated genes in the A. japonicus transcriptome (Supplementary Table S2). To enhance stability, the siRNA sequences were modified with 2’-O-methylation. Sea cucumbers that received no treatment were used as an additional blank negative control (NC) to eliminate any injection effects. These siRNAs were dissolved in diethyl pyrocarbonate (DEPC)-treated seawater to a final concentration of 20 μmol/L. Working solutions were prepared by mixing 20 μL of siRNA, 20 μL of Lipo6000™ transfection reagent (Beyotime, China), and 60 μL of RNAase-free seawater. Each group consisted of 10 sea cucumbers (approximately 30 g), which were injected with 0.1% (v/m) of the working solution, forming both experimental and control groups. Twenty-four hours after injection, CNR tissues were collected to evaluate the knockdown efficiency of the two siRNA_AjTRHP using qPCR. The siRNA_AjTRHP with superior performance (knockdown rate>50%) was selected for further experiments. For subsequent treatments, siRNAs were injected into the coelom via the tentacles every 24 h over a five-day period. Following the treatment phase, CNR and intestinal tissues from six individuals were dissected and frozen in liquid nitrogen and analyzed for the relative expression of AjCCKP1 (GenBank accession No. MH636358).
Statistical analyses for qPCR experiments were conducted using one-way analysis of variance (ANOVA) with Tukey post-hoc tests in SPSS v.27 (SPSS, USA). For the culture experiments, linear mixed models were applied to account for tank effects as random factors, with significance assessed through Tukey post-hoc tests conducted in R v.4.4.0.
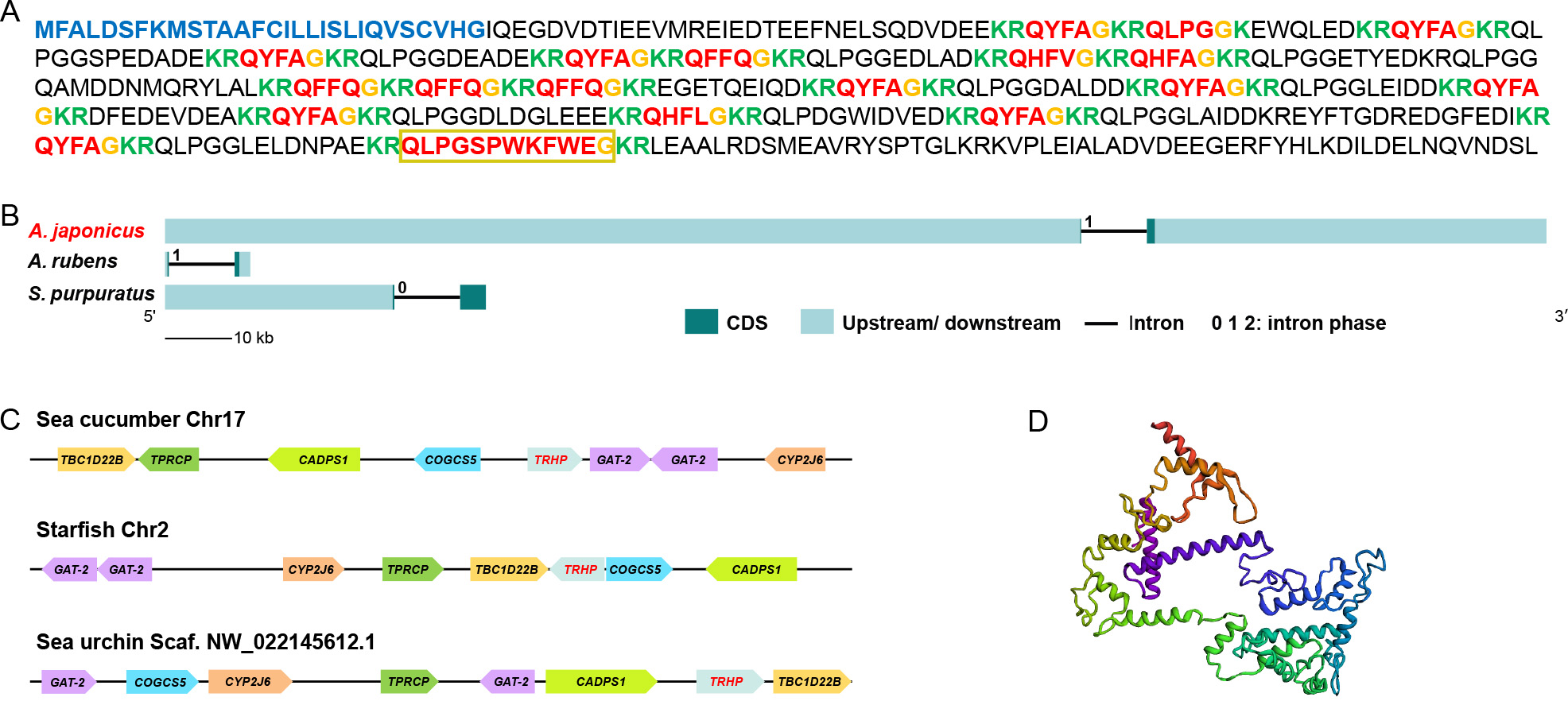
The sequences of AjTRH precursors are shown in Figure 1A. Gene structure analysis revealed that the TRH precursor proteins in sea cucumbers, starfish, and sea urchins were all encoded by two exons and one intron, with intron phases primarily annotated as “0” or “1” (Figure 1B). A conserved TRH superfamily domain was predicted across these three groups (Supplementary Figure S1). The genomic context of AjTRHP was assessed in the reference genome, revealing neighboring genes such as TBC1 domain family member 22B isoform X1 (TBC1D22B), TPR repeat-containing protein DDB_G0287407-like (TPRCP), calcium-dependent secretion activator 1 (CADPS1), putative conserved oligomeric Golgi complex subunit 5 (COGCS5), two copies of putative sodium- and chloride-dependent GABA transporter 2 (GAT-2), and putative cytochrome P450 2J6 (CYP2J6) (Figure 1C). The location of neighboring ortholog genes was also examined in other clades of echinoderms, using A. rubens (GenBank accession number: GCA_902459465.3) and S. purpuratus (GenBank accession number: GCA_000002235.4). Results showed that these synteny blocks were also located around the TRHP gene (Figure 1C), indicating a conserved synteny encompassing the TRH region during echinoderm evolution.
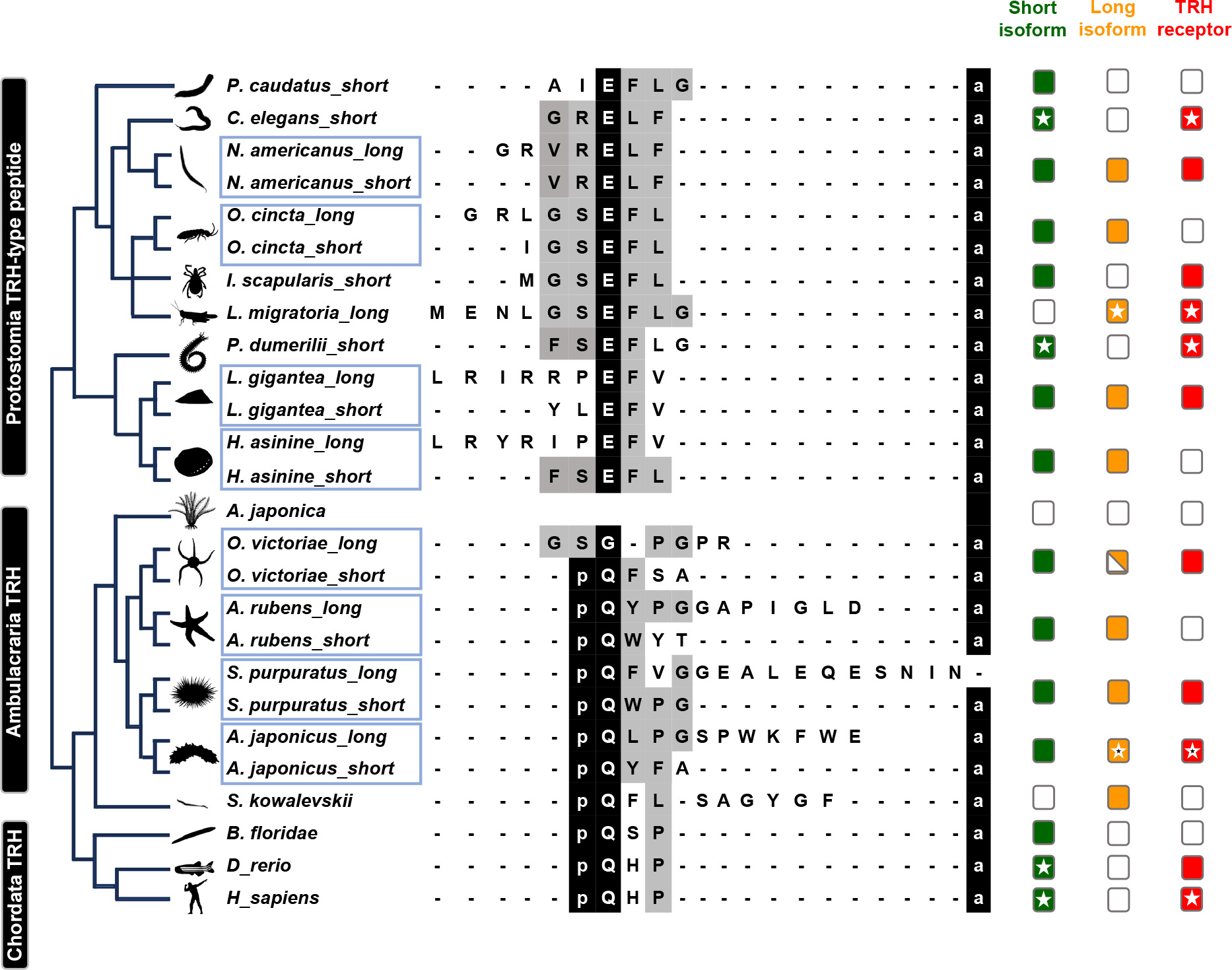
The molecular weight of the AjTRHP protein was predicted to be 52.54 kDa (Figure 1D; 3D-model). C-terminal amide modification emerged as a conserved feature among bilaterians, except for the long-isoform peptides of the echinoderm S. purpuratus. Notably, N-terminal modifications were conserved only within the deuterostomian clade. Long isoforms of the TRH/TRH-type peptides were only identified in protostomes (TRH-type peptides exceeding six amino acids) and Ambulacraria (TRH peptides exceeding four amino acids). However, TRH signaling components have not been identified in crinoid species to date (Figure 2).
The putative A. japonicus TRH receptor gene (AjTRHR) was identified based on in silico analysis of genomic and transcriptomic data. After cDNA cloning, the full-length cDNA of candidate AjTRHR (GenBank accession number: WLK66377.1) was obtained and characterized, spanning 1 823 bp with an ORF of 1 110 bp. Structural analysis using the Protter program revealed the presence of seven TMDs with an extracellular N-terminus, a cytoplasmic C-terminus, and typical glycosylation modifications (Supplementary Figure S2A). The 3D structural model of the candidate receptor is displayed in Supplementary Figure S2B. Phylogenetic analysis indicated that three receptor families were well grouped into three clades, with TRHR exhibiting a closer phylogenetic relationship with KissR than GnRHR. Within the TRHR clade, the candidate AjTRHR clustered with its counterpart in S. purpuratus and subsequently clustered with the ophiuroid species O. victoriae. These echinoderm TRHRs formed a sister group to TRHRs from other deuterostomes, which were clearly distinct from the protostomian clade (Figure 3).
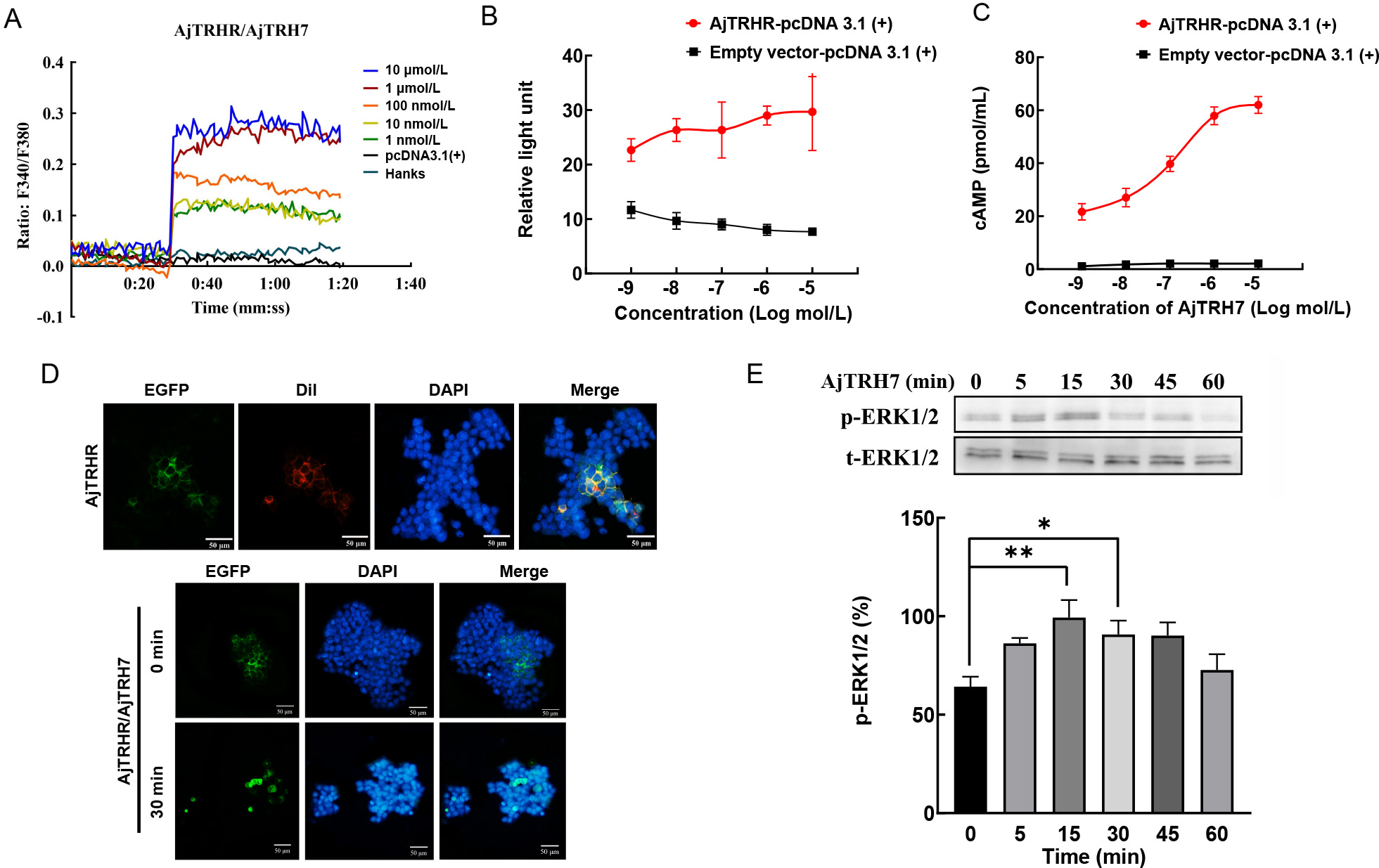
As shown in Figure 4A–C, increasing concentrations of synthesized AjTRH7 (10−9–10−5 mol/L) triggered intracellular Ca2+ mobilization (Figure 4A) and cAMP accumulation. The CRE-Luc reporter gene assay demonstrated that AjTRH7 activated AjTRHR (GenBank ID: OQ943938) signaling via the cAMP-PKA pathway (Figure 4B, C). Fluorescent cell staining revealed that AjTRHR expression was predominantly localized at the cell membrane (Figure 4D). An agonist-mediated receptor internalization assay, conducted in HEK293T cell expressing AjTRHR-pEGFP-N1, showed significant receptor internalization following stimulation with AjTRH7 for 30 min (Figure 4D). Results also indicated that p-ERK1/2, normalized to total ERK1/2 (t-ERK1/2), was gradually up-regulated, peaking 15 min after treatment with AjTRH7 and subsequently returning to baseline levels. Notably, p-ERK1/2 activity was markedly increased at 15 and 30 min following AjTRH7 stimulation (Figure 4E; F(5, 10)=5.563, P<0.05), confirming that AjTRHR is activated by AjTRH7 via the ERK1/2 phosphorylation pathway within the MAPK signaling cascade. Control experiments using the empty pcDNA 3.1(+) vector showed no significant responses (Figure 4E). Similarly, tests with reverse peptide sequences (EWFKWPSGPLQ) yielded no observable responses (Supplementary Figure S3B, C).
The relative expression levels of AjTRHP in different tissues were examined by qPCR, revealing that AjTRHP was mainly expressed in CNR tissue (Supplementary Figure S4A). Furthermore, ISH analysis provided a detailed localization profile of AjTRHP, as shown in Supplementary Figure S4B–H. Positive signals were observed at the margins and within the connective tissue linking the radial nerve cord (RNC) and CNR (Supplementary Figure S4B, C). Additionally, AjTRHP-positive staining was identified in the connective tissues of the intestine (Supplementary Figure S4D–F).
The localization of AjTRHR transcripts was consistent with that of AjTRHP. The ISH results demonstrated that AjTRHR was primarily expressed in the RNC of the nervous system (Supplementary Figure S5A–C). Positive signals were also detected in the connective tissues of the intestine but not in the intestinal lumen (Supplementary Figure S5D–G). Negative control experiments using sense probes confirmed the specificity of the observed staining patterns (Supplementary Figure S4G, H; Supplementary Figure S5H, I). Notably, as reported in our previous study on A. japonicus (Li et al., 2022), the simple columnar epithelium of the intestine was prone to non-specific staining with sense probes.
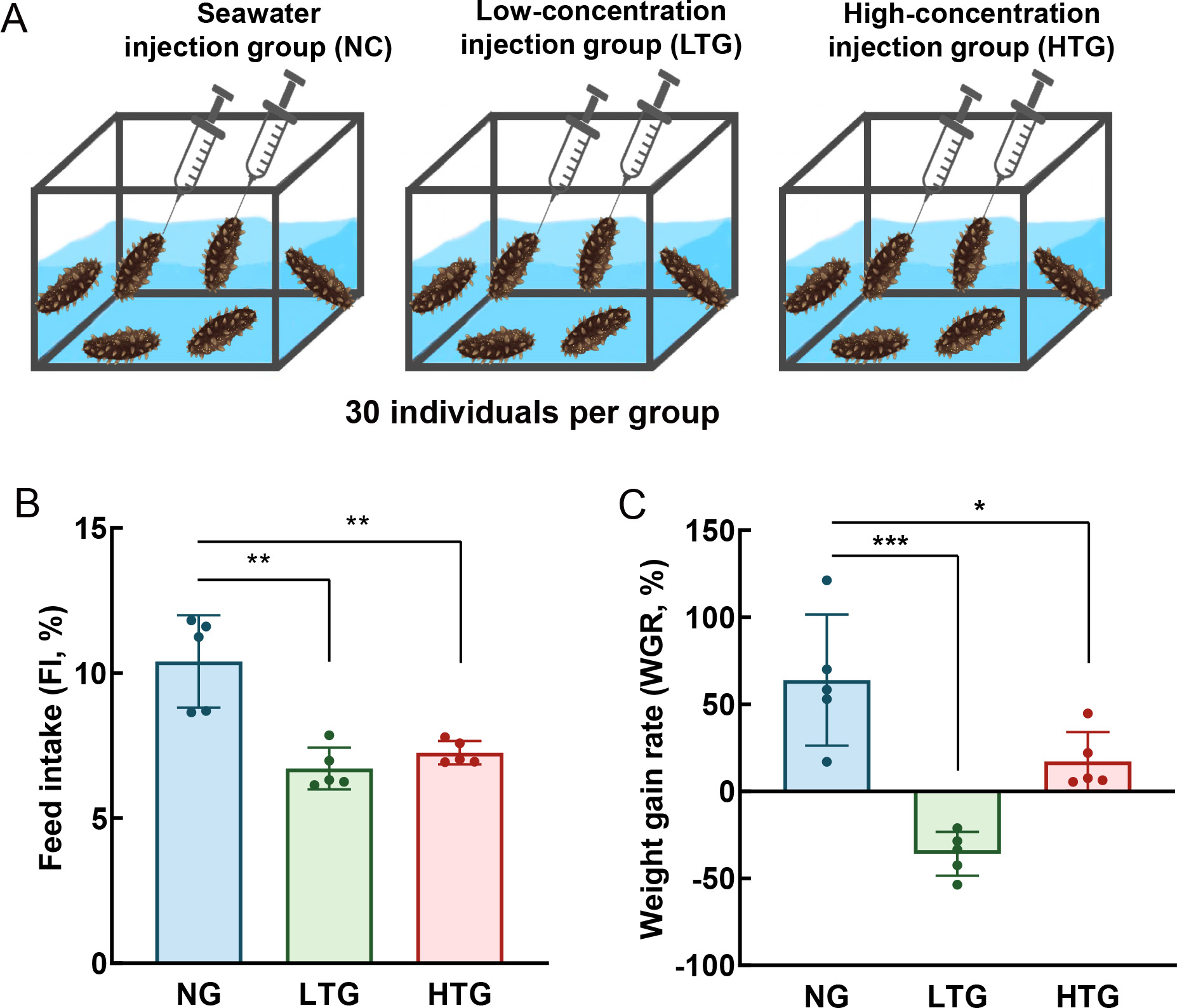
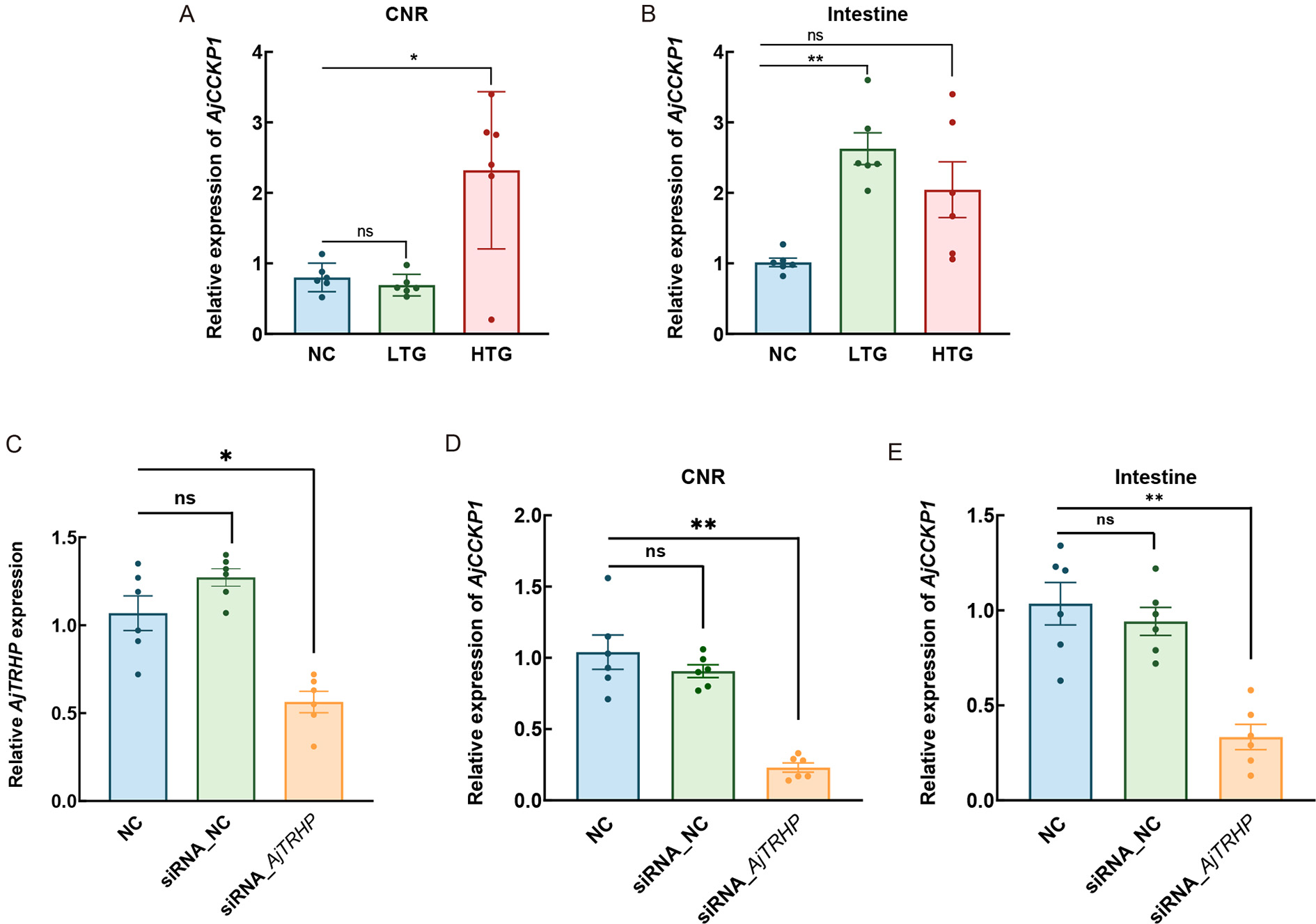
To explore the physiological role of TRH-type peptides in A. japonicus, experimental animals were exposed to AjTRH7 at two concentrations (0.5 mg/mL and 0.05 mg/mL). Injections were administered through the mouth into the coelom every two days over a one-month period. Ninety individuals were randomly divided into three groups: seawater injection group (NC), low-concentration AjTRH7 injection group (LTG), and high-concentration injection group (HTG), as displayed in Figure 5A. Results demonstrated significant effects of AjTRH7 on the WGR (F(2, 8)=5.248, P<0.05) and FI (F(2, 8)=18.509, P<0.05) of sea cucumbers. Sea cucumbers in the LTG and HTG groups exhibited significantly lower WGR compared to those in the NC group (P<0.05) (Figure 5B). Interestingly, AjTRH7 exhibited a stronger inhibitory effect in the LTG group than in the HTG group. Similarly, FI was significantly reduced in both the LTG and HTG groups compared to the NC group (Figure 5C).
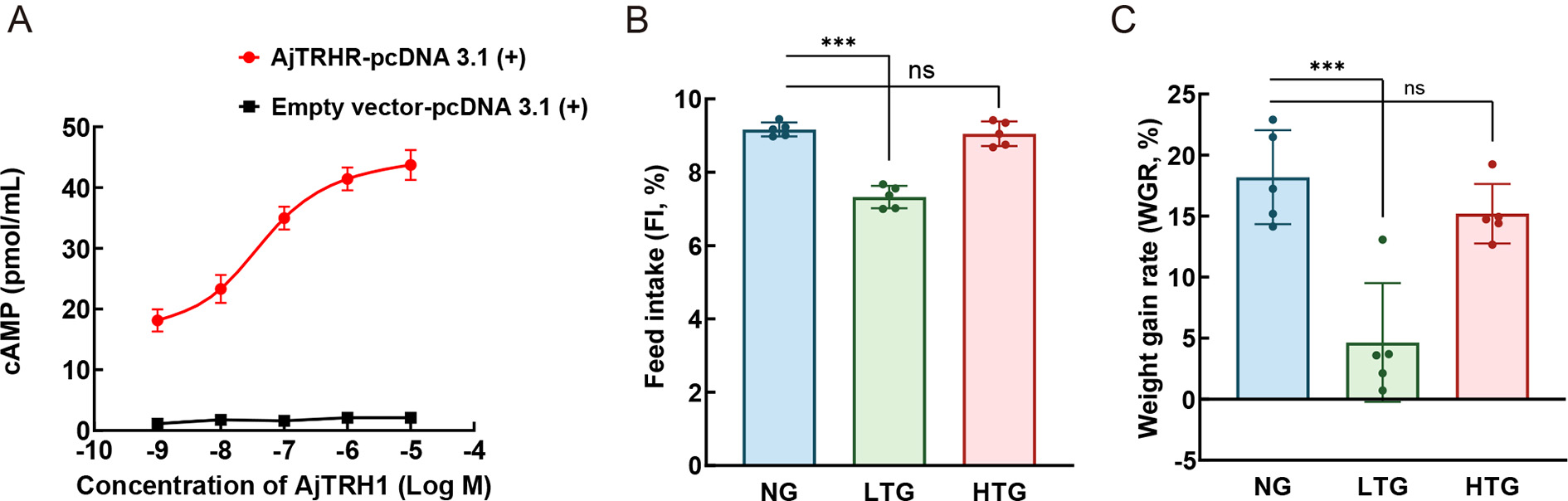
The short-isoform peptide AjTRH1 also binds to AjTRHR, the receptor targeted by AjTRH7, leading to cAMP accumulation (Figure 6A). However, unlike AjTRH7, no Ca2+ mobilization was observed after AjTRH1 stimulation (Supplementary Figure S6). Control experiments with peptides containing reversed sequences (AFYQ) also showed no responses (Supplementary Figure S3A). Pharmacological experiments revealed that low-concentration (0.05 mg/mL) AjTRH1 (LTG) significantly inhibited FI (F(2, 8)=65.748, P<0.05) and reduced WGR (F(2, 8)=21.666, P<0.05). In contrast, high-concentration (0.5 mg/mL) AjTRH1 (HTG) did not produce significant differences in WGR or FI compared to the NC group (Figure 6B, C).
Results demonstrated significant differences in AjCCKP1 expression among the three groups under AjTRH7 treatment in the CNR (F(2, 10)=6.852, P<0.05) and intestine (F(2, 10)=8.091, P<0.05). Expression levels of AjCCKP1 were significantly up-regulated in the CNR of the HTG group and in the intestines of the LTG group (Figure 7A, B). To further investigate this interaction, specific siRNA targeting AjTRHP (siRNA_AjTRHP) was applied to knockdown its expression (Figure 7C–E), resulting in a 55% reduction compared to the NC group (Figure 7C). Following five days of siRNA transfection (daily injections), the expression of AjCCKP1 mRNA was significantly inhibited in the CNR (F(2, 10)=16.875, P<0.05) and intestine (F(2, 10)=10.744, P<0.05) (Figure 7D, E).
Food intake is tightly regulated by the integration of internal and external environmental signals processed through neural circuits (Pool & Scott, 2014). In mammals, TRH signaling has been shown to play a critical role in feeding behavior by interacting with the satiety peptide CCK (Akieda-Asai et al., 2014; Wang et al., 2022). However, whether this interaction represents an evolutionarily conserved mechanism underlying feeding regulation in non-vertebrate animals has remained largely unexplored. This study provides the first evidence of functional responses elicited by a TRH-like peptide acting through a TRHR-like receptor in an invertebrate deuterostome, specifically the sea cucumber A. japonicus. Our findings elucidated the physiological role of TRH-like signaling in feeding and its interaction with the satiety peptide CCK, underscoring the conserved nature of this mechanism across evolutionary lineages.
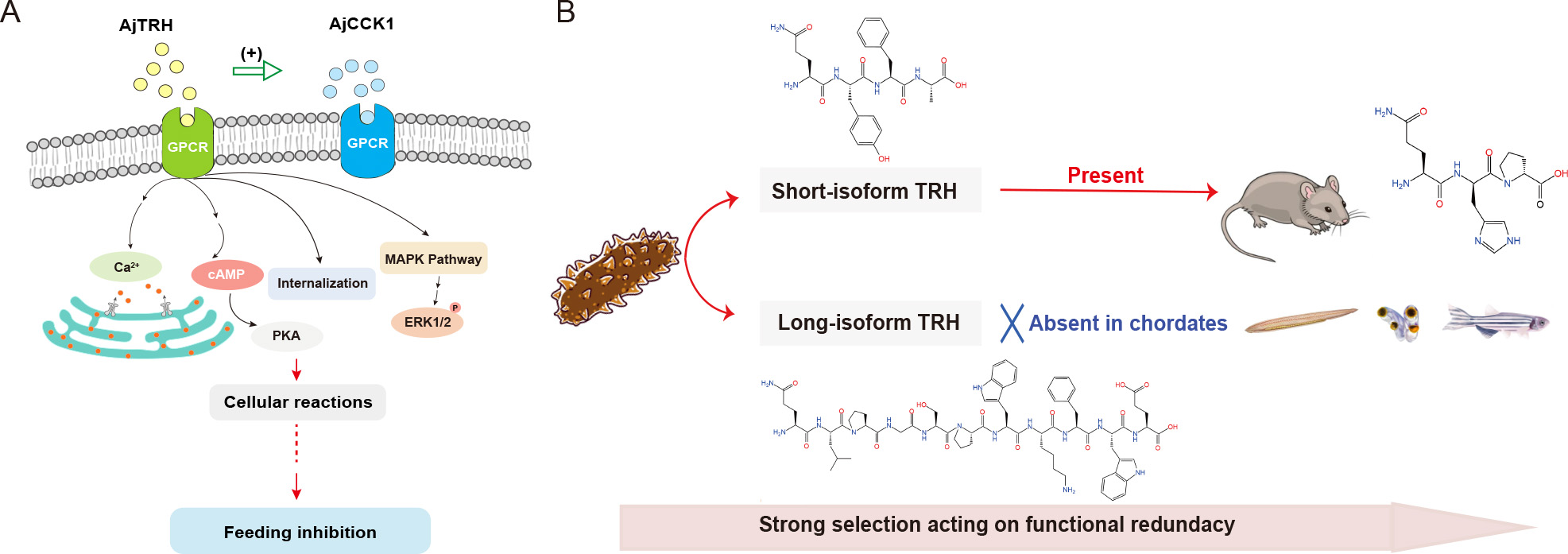
AjTRH signaling, like most neuropeptide systems, is mediated through G protein-coupled receptors (GPCRs) known as TRHRs. In vertebrates, TRHRs are well-classified as Class A peptide GPCRs, typically signaling through Gq/11 proteins but also capable of activating other G protein subtypes under certain conditions (Ji et al., 2022; Trubacova et al., 2022). For example, activated TRHR couples to Gq to trigger the phosphatidylinositol (IP3)-calcium-protein kinase C (PKC) pathway (Xu et al., 2022), which subsequently activates the Ca2+/CAMK (calmodulin-dependent protein kinase) and MAPK cascades (Cui et al., 1994; Trubacova et al., 2022). TRH-mediated signaling demonstrates diverse functional outcomes across species. For instance, TRH transiently induces intercellular Ca2+ influx in rat vagal nodose ganglion neurons, a response shorter in duration compared to other neuropeptides such as CCK-8, 5-HT, and ATP (Mamedova et al., 2022). In the frog species Rana ridibunda, TRHR activation mobilizes intracellular Ca2+ stores via Phospholipase C (PLC) activity (Galas et al., 1998). Similarly, TRH treatment in mice has been shown to induce significant changes in endoplasmic reticulum (ER) Ca2+ levels ([Ca2+]ER) in the pituitary gland (Rojo-Ruiz et al., 2021). Furthermore, ERK/MAPK pathway activity has been reported to regulate TRHR expression in Sprague-Dawley rats, increasing under pathway activation and decreasing with its inhibition (Liu et al., 2015). In this study, we identified a functional TRH receptor (AjTRHR) in the sea cucumber A. japonicus, marking the first experimental demonstration of TRHR activity in non-chordate deuterostomes. Using bioinformatics and ligand-binding assays, we confirmed that synthetic AjTRH7 acts as an agonist for AjTRHR, functioning via a cAMP/PKA-ERK/MAPK cascade and possibly via Ca2+-dependent pathway. AjTRH1, the short-isoform peptide, also activates AjTRHR by increasing cAMP levels, though it does not induce significant Ca2+ influx. Neuropeptide signaling systems are inherently complex, often involving multiple GPCR subtypes that linked to multiple signaling cascades. A single receptor can exhibit functional versatility, as different ligands binding to the same receptor may activate distinct G protein subtypes within the same cell (Mendel et al., 2020; Møller et al., 2003). The functional diversity is influenced by receptor structural polymorphism and variation in ligand-binding sites (Changeux, 2010; Wong et al., 2000), suggestingthat AjTRH1 and AjTRH7 may engage distinct binding domains on AjTRHR. Furthermore, it is plausible that additional, yet unidentified, receptors may mediate the activity of AjTRH peptides, contributing to the observed complexity of this signaling system.
Interestingly, relative expression of AjTRHP in the intestine of A. japonicus has been shown to increase significantly during the deep stages of aestivation, a state characterized by metabolic arrest and feeding cessation (Zhao et al., 2014). These observations, combined with findings from the current study, suggest a functional role for TRH in feeding inhibition during sea cucumber aestivation. This role aligns with findings in hibernating mammals, where endogenous TRH plays a crucial role in hypothermic states by reducing food intake and elevating body temperature, as observed in hamsters (Tamura et al., 2005). Similarly, increased TRH mRNA expression and activation of TRH neurons have been documented in torpor thirteen-lined ground squirrels (Ictidomys tridecemlineatus) and Arctic ground squirrel (Urocitellus parryii), respectively (Frare et al., 2021; Schwartz et al., 2013). Therefore, we speculate that TRH may play a broader role in seasonal animal dormancy, such as aestivation, hibernation, and torpor, particularly in the regulation of feeding behavior during metabolic arrest. However, further research is needed to clarify the role of TRH signaling in these processes, providing deeper insights into its mechanisms and advancing research on metabolic arrest in animals.
In vivo pharmacological experiments demonstrated that TRH-type neuropeptides significantly suppressed food intake, ultimately leading to body mass reduction in A. japonicus. Interestingly, this inhibitory effect was more pronounced at a lower concentration (0.05 mg/mL) compared to a higher concentration (0.5 mg/mL), possibly due to negative feedback mechanisms regulating endogenous TRH secretion.
In mammals, the interaction between TRH and CCK has been shown to modulate satiety signals, effectively reducing food consumption (Akieda-Asai et al., 2014; Wang et al., 2022). Similarly, in the echinoderm A. rubens, SK/CCK-type peptides have been implicated in suppressing the onset of feeding (Tinoco et al., 2021). To explore whether this TRH-CCK interaction is conserved in invertebrates, we explored its role in feeding regulation in A. japonicus. Experimental evidence showed that AjCCK1.2 significantly inhibited feeding in sea cucumbers (unpublished data). Notably, the relative expression of AjCCKP1 closely mirrored the expression patterns of AjTRH7, increasing with AjTRH7 overexpression and decreasing following AjTRHP knockdown. These findings suggest that TRH interacts with appetite-regulating peptides such as CCK to inhibit feeding behavior in A. japonicus (Figure 8A). Additionally, the expression and localization patterns of AjTRHP in the nervous system and intestine further supported its pivotal role in feeding regulation in sea cucumbers.
Feeding behaviors are hypothesized to be under complex control of regulatory networks involving the synergistic actions of multiple neuropeptides (Gotoh et al., 2013; Guo et al., 2021; Loh et al., 2017; Matson et al., 2000; Nässel & Wu, 2022; Peters et al., 2006; Zels et al., 2015). In mammals, TRH is widely classified as an anorectic factor. For instance, prolonged TRH administration has been shown to elevate resting metabolic rates and suppress food intake, leading to reduced body weight in obese rats (Al-Arabi & Andrews, 2006). Central administration of TRH has similarly been found to decrease food intake in Sprague-Dawley rats and Siberian hamsters (Phodopus sungorus) (Choi et al., 2002; Schuhler et al., 2007; Steward et al., 2003). The regulation of feeding behavior in mammals is mediated by both central and peripheral mechanisms. Central regulation involves TRH production in second-order neurons located in the paraventricular nucleus (PVN) of the hypothalamus, while peripheral regulation integrates signals from CCK and other factors that communicate with the brain to modulate central responses, thus regulating feeding behavior (Valassi et al., 2008). Recent structural analyses of TRHR have revealed striking similarities to the CCK A receptor (CCKAR) (Ji et al., 2022), providing a molecular basis for the interaction between TRH and CCK signaling pathways. In this study, we demonstrated that TRH stimulates the release of the satiety peptide CCK, reducing food intake via satiety signaling. This finding underscores the conserved role of TRH-CCK interactions in regulating feeding behavior. However, in echinoderms, the integrated regulatory networks and neuronal circuits controlling feeding behavior remain largely unknown, limiting our understanding of the function and evolution of feeding-related peptidergic signals in these animals. Further research is essential to elucidate the mechanisms underlying the interactions between TRH, CCK, and other feeding-related peptides. Such studies will provide critical insights into the molecular pathways governing feeding behavior, particularly in marine invertebrates, and expand our understanding of the evolutionary dynamics of neuropeptide signaling in animals.
The sequence of TRH (tripeptide pQHP-NH2) is fully conserved across vertebrates, indicating strong evolutionary pressure to maintain its structure and physiological functions. However, in invertebrate deuterostomes, such as echinoderms, two distinct types of TRH peptides have been identified: short and long isoforms (Figure 8B). The short peptides (tetrapeptide) are considered orthologs of vertebrate TRH (Van Sinay et al., 2017). In contrast, the long peptides exhibit unique structural and functional characteristics, suggesting a distinct evolutionary trajectory in which they were retained specifically in Ambulacraria. Our functional experiments demonstrated that both short (AjTRH1) and long isoforms (AjTRH7) inhibited food intake in sea cucumbers, indicating functional conservation of the short isoform with vertebrate TRH. Interestingly, in protostomes, EFLGa has been identified as the ligand for the TRH receptor ortholog in Platynereis, sharing some similarity with the TRH ortholog in the sea urchin Strongylocentrotus purpuratus. This finding suggests an evolutionary relationship between EFLGa and deuterostome TRH (Bauknecht & Jékely, 2015; Rowe & Elphick, 2012). Based on these similarities, we classified these peptides collectively as “TRH-type”. TRH-type neuropeptides have been identified in other protostomes (Van Sinay et al., 2017; Veenstra & Šimo, 2020; Zandawala et al., 2024), including Necator americanus, Orchesella cincta, Lottia gigantea, and Haliotis asinina, which contain both short and long isoforms. The absence of long isoforms in vertebratesmay reflect natural selection acting on functional redundancy between long and short isoforms. Alternatively, the long isoform may have arisen independently in the ambulacrarian ancestor, suggesting a more parsimonious evolutionary scenario. From a phylogenetic perspective, the functional redundancy of the two isoforms is plausible, as both can independently regulate feeding behavior. However, variations in the strength of their inhibitory effects have been observed, with the long isoform functioning through two pathways, while the short isoform acts via the cAMP pathway. These differences may arise from distinct signaling pathways or species-specific ecological contexts, although there is overlap in the way both isoforms function. Further research is required to determine whether organisms with only the short isoform exhibit lower feeding inhibition compared to those with the long isoform, or whether this variation is context-dependent, influenced by ecological or evolutionary pressures. Expanding studies across additional species will be critical to refine our understanding of the evolutionary history of TRH signaling and its functional adaptations.
This study advances our understanding of the function and signaling pathways of ancestral TRH-type neuropeptides, shedding light on their evolutionary and physiological significance. Our findings demonstrate that intercellular Ca2+ mobilization, receptor internalization, and cAMP accumulation are induced when TRH binds to its functional receptor, AjTRHR, which operates via the ERK1/2 phosphorylation pathway within the MAPK cascade. Furthermore, TRH signaling suppresses feeding activity by interacting with CCK-type neuropeptide-mediated satiety signals, a mechanism likely conserved in aestivating A. japonicus. The observed functional redundancy of TRH peptides suggests that the loss of long-isoform TRH in chordates reflects evolutionary trade-offs driven by natural selection. Differences in signaling mechanisms between long and short isoforms suggest potential context-dependent or nuanced roles that warrant further exploration. Future studies examining TRH signaling across diverse species will be crucial to confirm the proposed evolutionary history of TRH signaling. This work offers novel insights into the evolutionary history of TRH signaling and highlights the cooperative interactions of peptidergic pathways in regulating feeding behavior in marine invertebrates.
Supplementary data to this article can be found online.
The authors declare that they have no competing interests.
Conceptualization: M.C.; Methodology: Y.Z., M.C., and J.D.G.-E.; Data curation: Y.Z., H.L., and X.D.; Investigation: Y.Z., H.L., and X.D.; Writing–original draft: Y.Z.; Formal analysis: M.C. and J.D.G.-E.; Writing–review and editing: M.C. and J.D.G.-E.; Supervision: M.C.; Funding acquisition: M.C. All authors read and approved the final version of the manuscript.
|
Akieda-Asai S, Poleni PE, Date Y. 2014. Coinjection of CCK and leptin reduces food intake via increased CART/TRH and reduced AMPK phosphorylation in the hypothalamus. American Journal of Physiology-Endocrinology and Metabolism, 306(11): E1284–E1291. DOI: 10.1152/ajpendo.00664.2013
|
|
Al-Arabi A, Andrews JF. 2006. The effect of TRH and norepinephrine on the triglyceride droplets (TGD) in brown adipose tissue in warm acclimated rats. Biomedical Sciences Instrumentation, 42: 507–512.
|
|
Bauknecht P, Jékely G. 2015. Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. Cell Reports, 12(4): 684–693. DOI: 10.1016/j.celrep.2015.06.052
|
|
Burbach JPH. 2011. What are neuropeptides?. In: Merighi A. Neuropeptides. New York: Humana Press, 1–36.
|
|
Chaiyamoon A, Tinikul R, Nontunha N, et al. 2020. Characterization of TRH/GnRH-like peptides in the sea cucumber, Holothuria scabra, and their effects on oocyte maturation. Aquaculture, 518: 734814. DOI: 10.1016/j.aquaculture.2019.734814
|
|
Changeux JP. 2010. Allosteric receptors: from electric organ to cognition. Annual Review of Pharmacology and Toxicology, 50: 1–38. DOI: 10.1146/annurev.pharmtox.010909.105741
|
|
Chen MY, Talarovicova A, Zheng YQ, et al. 2019. Neuropeptide precursors and neuropeptides in the sea cucumber Apostichopus japonicus: a genomic, transcriptomic and proteomic analysis. Scientific Reports, 9(1): 8829. DOI: 10.1038/s41598-019-45271-3
|
|
Chieu HD, Suwansa-Ard S, Wang TF, et al. 2019. Identification of neuropeptides in the sea cucumber Holothuria leucospilota. General and Comparative Endocrinology, 283: 113229. DOI: 10.1016/j.ygcen.2019.113229
|
|
Choi YH, Hartzell D, Azain MJ, et al. 2002. TRH decreases food intake and increases water intake and body temperature in rats. Physiology & Behavior, 77(1): 1–4.
|
|
Conzelmann M, Williams EA, Krug K, et al. 2013. The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genomics, 14: 906. DOI: 10.1186/1471-2164-14-906
|
|
Cui ZJ, Gorelick FS, Dannies PS. 1994. Calcium/calmodulin-dependent protein kinase-II activation in rat pituitary cells in the presence of thyrotropin-releasing hormone and dopamine. Endocrinology, 134(5): 2245–2250. DOI: 10.1210/endo.134.5.8156928
|
|
Frare C, Williams CT, Drew KL. 2021. Thermoregulation in hibernating mammals: the role of the “thyroid hormones system”. Molecular and Cellular Endocrinology, 519: 111054. DOI: 10.1016/j.mce.2020.111054
|
|
Galas L, Lamacz M, Garnier M, et al. 1998. Involvement of extracellular and intracellular calcium sources in TRH-induced α-MSH secretion from frog melanotrope cells. Molecular and Cellular Endocrinology, 138(1-2): 25–39. DOI: 10.1016/S0303-7207(98)00053-7
|
|
Gotoh K, Masaki T, Chiba S, et al. 2013. Nesfatin‐1, corticotropin‐releasing hormone, thyrotropin‐releasing hormone, and neuronal histamine interact in the hypothalamus to regulate feeding behavior. Journal of Neurochemistry, 124(1): 90–99. DOI: 10.1111/jnc.12066
|
|
Guo D, Zhang YJ, Zhang S, et al. 2021. Cholecystokinin-like peptide mediates satiety by inhibiting sugar attraction. PLoS Genetics, 17(8): e1009724. DOI: 10.1371/journal.pgen.1009724
|
|
Huang BW, Lv ZM, Li YN, et al. 2021. Identification and functional characterization of natural resistance-associated macrophage protein 2 from sea cucumber Apostichopus japonicus. Developmental & Comparative Immunology, 114: 103835.
|
|
Jékely G. 2013. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proceedings of the National Academy of Sciences of the United States of America, 110(21): 8702–8707.
|
|
Ji SY, Dong YJ, Chen LN, et al. 2022. Molecular basis for the activation of thyrotropin-releasing hormone receptor. Cell Discovery, 8(1): 116. DOI: 10.1038/s41421-022-00477-0
|
|
Li CY, Zheng YQ, Cong X, et al. 2022. Molecular and functional characterization of the luqin-type neuropeptide signaling system in the sea cucumber Apostichopus japonicus. Peptides, 155: 170839. DOI: 10.1016/j.peptides.2022.170839
|
|
Li G, Shi Y, Huang HS, et al. 2010. Internalization of the human nicotinic acid receptor GPR109A is regulated by Gi, GRK2, and arrestin3. Journal of Biological Chemistry, 285(29): 22605–22618. DOI: 10.1074/jbc.M109.087213
|
|
Liu CJ, Li LB, Ha M, et al. 2015. The PI3K/Akt and ERK pathways elevate thyroid hormone receptor β1 and TRH receptor to decrease thyroid hormones after exposure to PCB153 and p, p′-DDE. Chemosphere, 118: 229–238. DOI: 10.1016/j.chemosphere.2014.09.023
|
|
Liu YZ, He G, Wang QC, et al. 2014. Hydroxyproline supplementation on the performances of high plant protein source based diets in turbot (Scophthalmus maximus L. ). Aquaculture, 433: 476–480. DOI: 10.1016/j.aquaculture.2014.07.002
|
|
Loh K, Zhang L, Brandon A, et al. 2017. Insulin controls food intake and energy balance via NPY neurons. Molecular Metabolism, 6(6): 574–584. DOI: 10.1016/j.molmet.2017.03.013
|
|
Mamedova E, Dmytriyeva O, Rekling JC. 2022. Thyrotropin-releasing hormone induces Ca2+ increase in a subset of vagal nodose ganglion neurons. Neuropeptides, 94: 102261. DOI: 10.1016/j.npep.2022.102261
|
|
Matson CA, Reid DF, Cannon TA, et al. 2000. Cholecystokinin and leptin act synergistically to reduce body weight. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology, 278 (4): R882–R890.
|
|
Mayorova TD, Tian S, Cai WG, et al. 2016. Localization of neuropeptide gene expression in larvae of an echinoderm, the starfish Asterias rubens. Frontiers in Neuroscience, 10: 553.
|
|
Mendel HC, Kaas Q, Muttenthaler M. 2020. Neuropeptide signalling systems–An underexplored target for venom drug discovery. Biochemical Pharmacology, 181: 114129. DOI: 10.1016/j.bcp.2020.114129
|
|
Møller LN, Stidsen CE, Hartmann B, et al. 2003. Somatostatin receptors. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1616(1): 1–84. DOI: 10.1016/S0005-2736(03)00235-9
|
|
Nässel DR, Wu SF. 2022. Cholecystokinin/sulfakinin peptide signaling: conserved roles at the intersection between feeding, mating and aggression. Cellular and Molecular Life Sciences, 79(3): 188. DOI: 10.1007/s00018-022-04214-4
|
|
Ohta N, Kubota I, Takao T, et al. 1991. Fulicin, a novel neuropeptide containing a D-amino acid residue isolated from the ganglia of Achatina fulica. Biochemical and Biophysical Research Communications, 178(2): 486–493. DOI: 10.1016/0006-291X(91)90133-R
|
|
Peters JH, Simasko SM, Ritter RC. 2006. Modulation of vagal afferent excitation and reduction of food intake by leptin and cholecystokinin. Physiology & Behavior, 89(4): 477–485.
|
|
Pool AH, Scott K. 2014. Feeding regulation in Drosophila. Current Opinion in Neurobiology, 29: 57–63. DOI: 10.1016/j.conb.2014.05.008
|
|
Puga L, Alcántara-Alonso V, Coffeen U, et al. 2016. TRH injected into the nucleus accumbens shell releases dopamine and reduces feeding motivation in rats. Behavioural Brain Research, 306: 128–136. DOI: 10.1016/j.bbr.2016.03.031
|
|
Rojo-Ruiz J, Navas-Navarro P, Nuñez L, et al. 2021. Imaging of endoplasmic reticulum Ca2+ in the intact pituitary gland of transgenic mice expressing a low affinity Ca2+ indicator. Frontiers in Endocrinology, 11: 615777. DOI: 10.3389/fendo.2020.615777
|
|
Root CM, Ko KI, Jafari A, et al. 2011. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell, 145(1): 133–144. DOI: 10.1016/j.cell.2011.02.008
|
|
Rowe ML, Achhala S, Elphick MR. 2014. Neuropeptides and polypeptide hormones in echinoderms: new insights from analysis of the transcriptome of the sea cucumber Apostichopus japonicus. General and Comparative Endocrinology, 197: 43–55. DOI: 10.1016/j.ygcen.2013.12.002
|
|
Rowe ML, Elphick MR. 2012. The neuropeptide transcriptome of a model echinoderm, the sea urchin Strongylocentrotus purpuratus. General and Comparative Endocrinology, 179(3): 331–344. DOI: 10.1016/j.ygcen.2012.09.009
|
|
Schuhler S, Warner A, Finney N, et al. 2007. Thyrotrophin‐releasing hormone decreases feeding and increases body temperature, activity and oxygen consumption in Siberian hamsters. Journal of Neuroendocrinology, 19(4): 239–249. DOI: 10.1111/j.1365-2826.2006.01524.x
|
|
Schwartz C, Hampton M, Andrews MT. 2013. Seasonal and regional differences in gene expression in the brain of a hibernating mammal. PLoS One, 8(3): e58427. DOI: 10.1371/journal.pone.0058427
|
|
Semmens DC, Mirabeau O, Moghul I, et al. 2016. Transcriptomic identification of starfish neuropeptide precursors yields new insights into neuropeptide evolution. Open Biology, 6(2): 150224. DOI: 10.1098/rsob.150224
|
|
Smith MK, Wang TF, Suwansa-Ard S, et al. 2017. The neuropeptidome of the Crown-of-Thorns Starfish, Acanthaster planci. Journal of Proteomics, 165: 61–68. DOI: 10.1016/j.jprot.2017.05.026
|
|
Steward CA, Horan TL, Schuhler S, et al. 2003. Central administration of thyrotropin releasing hormone (TRH) and related peptides inhibits feeding behavior in the Siberian hamster. Neuroreport, 14(5): 687–691. DOI: 10.1097/00001756-200304150-00006
|
|
Suwansa-Ard S, Chaiyamoon A, Talarovicova A, et al. 2018. Transcriptomic discovery and comparative analysis of neuropeptide precursors in sea cucumbers (Holothuroidea). Peptides, 99: 231–240. DOI: 10.1016/j.peptides.2017.10.008
|
|
Tamura Y, Shintani M, Nakamura A, et al. 2005. Phase-specific central regulatory systems of hibernation in Syrian hamsters. Brain research, 1045(1-2): 88–96. DOI: 10.1016/j.brainres.2005.03.029
|
|
Tinoco AB, Barreiro-Iglesias A, Guerra LAY, et al. 2021. Ancient role of sulfakinin/cholecystokinin-type signalling in inhibitory regulation of feeding processes revealed in an echinoderm. eLife, 10: e65667. DOI: 10.7554/eLife.65667
|
|
Trubacova R, Drastichova Z, Novotny J. 2022. Biochemical and physiological insights into TRH receptor-mediated signaling. Frontiers in Cell and Developmental Biology, 10: 981452. DOI: 10.3389/fcell.2022.981452
|
|
Valassi E, Scacchi M, Cavagnini F. 2008. Neuroendocrine control of food intake. Nutrition, Metabolism and Cardiovascular Diseases, 18 (2): 158–168.
|
|
Van Sinay E, Mirabeau O, Depuydt G, et al. 2017. Evolutionarily conserved TRH neuropeptide pathway regulates growth in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America, 114(20): E4065–E4074.
|
|
Veenstra JA, Šimo L. 2020. The TRH-ortholog EFLamide in the migratory locust. Insect Biochemistry and Molecular Biology, 116: 103281. DOI: 10.1016/j.ibmb.2019.103281
|
|
Wang TM, Cao Z, Shen ZF, et al. 2020. Existence and functions of a kisspeptin neuropeptide signaling system in a non-chordate deuterostome species. eLife, 9: e53370. DOI: 10.7554/eLife.53370
|
|
Wang TM, Yang Z, Zhou NM, et al. 2017. Identification and functional characterisation of 5-HT4 receptor in sea cucumber Apostichopus japonicus (Selenka). Scientific Reports, 7(1): 40247. DOI: 10.1038/srep40247
|
|
Wang YT, Xu WX, Zhang JM, et al. 2022. Effects of glycyrrhizin (GL) supplementation on survival, growth performance, expression of feeding-related genes, activities of digestive enzymes, antioxidant capacity, and expression of inflammatory factors in large yellow croaker (Larimichthys crocea) larvae. Aquaculture Nutrition, 2022: 5508120.
|
|
Wong AHC, Buckle CE, Van Tol HH. 2000. Polymorphisms in dopamine receptors: what do they tell us?. European Journal of Pharmacology, 410(2-3): 183–203. DOI: 10.1016/S0014-2999(00)00815-3
|
|
Xu Y W, Cai H M, You C Z, et al. 2022. Structural insights into ligand binding and activation of the human thyrotropin-releasing hormone receptor. Cell Research, 32(9): 855–857.
|
|
Zandawala M, Amir MB, Shin J, et al. 2024. Proteome-wide neuropeptide identification using NeuroPeptide-HMMer (NP-HMMer). General and Comparative Endocrinology, 357: 114597. DOI: 10.1016/j.ygcen.2024.114597
|
|
Zandawala M, Moghul I, Yañez Guerra LA, et al. 2017. Discovery of novel representatives of bilaterian neuropeptide families and reconstruction of neuropeptide precursor evolution in ophiuroid echinoderms. Open Biology, 7(9): 170129. DOI: 10.1098/rsob.170129
|
|
Zels S, Dillen S, Crabbé K, et al. 2015. Sulfakinin is an important regulator of digestive processes in the migratory locust, Locusta migratoria. Insect Biochemistry and Molecular Biology, 61: 8–16. DOI: 10.1016/j.ibmb.2015.03.008
|
|
Zhao Y, Yang HS, Storey KB, et al. 2014. RNA-seq dependent transcriptional analysis unveils gene expression profile in the intestine of sea cucumber Apostichopus japonicus during aestivation. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 10: 30–43. DOI: 10.1016/j.cbd.2014.02.002
|
|
Zheng YQ, Cong X, Liu HC, et al. 2022. Nervous system development and neuropeptides characterization in embryo and larva: insights from a non-chordate deuterostome, the sea cucumber Apostichopus japonicus. Biology, 11(10): 1538. DOI: 10.3390/biology11101538
|
|
Zheng YQ, Cong X, Liu HC, et al. 2024. Neuronal cell populations in circumoral nerve ring of sea cucumber Apostichopus japonicus: ultrastructure and transcriptional profile. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 52: 101263.
|
|
Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins. In Bryson V, Vogel HJ. Evolving Genes and Proteins. New York: Academic Press, 97–166.
|